Epigenomics
Principaux investigateurs

Prof. Stephan Frank
Leitender Arzt und Fachbereichsleiter Neuro- und Ophthalmopathologie
Pathologie
Tel. +41 61 328 63 90

Membres du groupe

Notre science
Nos efforts de recherche actuels se concentrent sur le diagnostic épigénomique des tumeurs basé sur l'apprentissage automatique. L'introduction d'un classificateur de la méthylation de l'ADN des tumeurs cérébrales (Capper et al., Nature 2018) a incité à mettre en place un pipeline de diagnostic correspondant dans notre institut. En utilisant des puces à ADN, nous avons jusqu'à présent analysé les profils de méthylation de l'ADN de plus de 2000 échantillons tumoraux dans un contexte de diagnostic. Toutefois, comme cette approche est chronophage, nous avons récemment commencé à mettre en place un pipeline basé sur le séquençage des nanopores pour permettre le profilage épigénomique (et génomique) le jour même. Son flux de travail sous-jacent (Euskirchen et al., Acta Neuropathol 2017) est beaucoup moins dépendant de l'équipement et des ressources, et permet le typage moléculaire des tumeurs avant même que les coupes histologiques ou les frottis spéciaux ne soient disponibles. En outre, nous venons de lancer un portail open-source permettant aux collègues du monde entier de comparer leurs jeux de données de méthylation avec un jeu de données de cas de référence annotés en constante augmentation(www.epidip.org). Enfin, en tant que membre d'un consortium international, nous avons contribué (et continuons à le faire) à un certain nombre d'efforts de collaboration ; publications pertinentes ci-dessous.
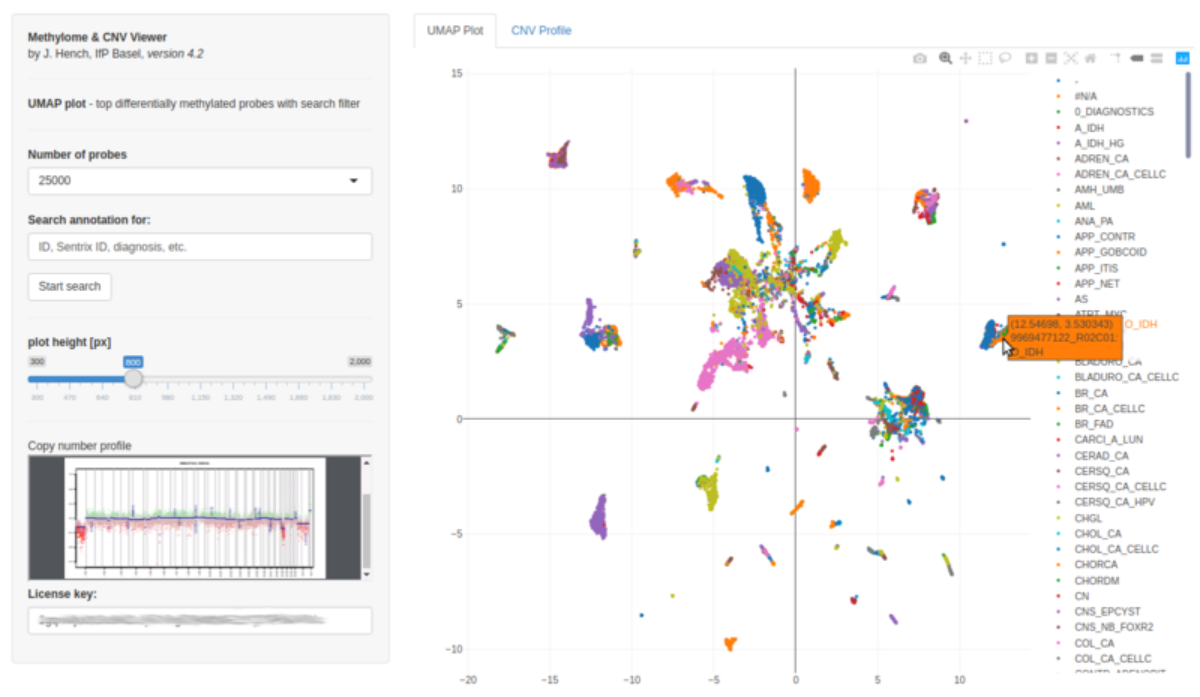
Capture d'écran de l'outil EpiDiP.
Publications sélectionnées
- Patel A, Dogan H, Payne A, et al. Rapid-CNS2: rapid comprehensive adaptive nanopore-sequencing of CNS tumors, a proof-of-concept study. Acta Neuropathol. 2022 Mar 31.
- Hench J, Vlajnic T, Soysal SD, et al. An Integrated Epigenomic and Genomic View on Phyllodes and Phyllodes-like Breast Tumors. Cancers (Bâle). 2022 Jan 28;14(3):667.
- Bockmayr M, Harnisch K, Pohl LC, et al. Comprehensive profiling of myxopapillary ependymomas identifies a distinct molecular subtype with relapsing disease . Neuro Oncol. 2022 Apr 5:noac088.
- Borlin PR, Brazzola P, Frontzek K, et al. Cancer chez les enfants présentant des variantes bialléliques de BRCA1 et des caractéristiques similaires à l'anémie de Fanconi : rapport d'une tumeur cérébrale maligne chez un jeune enfant. Pediatr Blood Cancer. 2022 avr 4:e29680.
- Maas SLN, Stichel D, Hielscher T, et al ; Consortium allemand sur les méningiomes agressifs (KAM). Integrated Molecular-Morphologic Meningioma Classification : A Multicenter Retrospective Analysis, Retrospectively and Prospectively Validated . J Clin Oncol. 2021 Oct 7:JCO2100784.
- Della Monica R, Cuomo M, Visconti R, et al. Evaluation of MGMT gene methylation in neuroendocrine neoplasms. Oncol Res. 2021 May 20
- Suwala AK, Stichel D, Schrimpf D, et al. Glioblastomas with primitive neuronal component harbor a distinct methylation and copy-number profile with inactivation of TP53, PTEN, and RB1. Acta Neuropathol. 2021 Jul;142(1):179-189.
- Glöss S, Jurmeister P, Thieme A, et al. IDH2 R172 Mutations Across Poorly Differentiated Sinonasal Tract Malignancies : Forty Molecularly Homogenous and Histologically Variable Cases With Favorable Outcome . Am J Surg Pathol. 2021 Sep 1;45(9):1190-1204.
- Hench IB, Monica RD, Chiariotti L, et al. Fast routine assessment of MGMT promoter methylation. Neurooncol Adv. 2020 Dec 5;3(1):vdaa170.
- Koelsche C, Schrimpf D, Stichel D, et al. Sarcoma classification by DNA methylation profiling. Nat Commun. 2021 Jan 21;12(1):498. doi : 10.1038/s41467-020-20603-4.
- Zschernack V, Jünger ST, Mynarek M, et al. Supratentorial ependymoma in childhood : more than just RELA or YAP. Acta Neuropathol. 2021 Jan 22. doi : 10.1007/s00401-020-02260-5.
- Sievers P, Sill M, Blume C, et al. Les méningiomes à cellules claires sont définis par un profil de méthylation de l'ADN très distinct et des mutations dans SMARCE1. Acta Neuropathol. 2020 Dec 14. doi : 10.1007/s00401-020-02247-2.
- Sievers P, Sill M, Schrimpf D, Stichel D, et al. A subset of pediatric-type thalamic gliomas share a distinct DNA methylation profile, H3K27me3 loss and frequent alteration of EGFR. Neuro Oncol. 2020 Nov 1:noaa251.
- Janin M, Ortiz-Barahona V, de Moura MC, et al. Epigenetic loss of RNA-methyltransferase NSUN5 in glioma targets ribosomes to drive a stress adaptive translational program. Acta Neuropathol. 2019 138(6):1053-1074. doi:10.1007/s00401-019-02062-4.
- Haefliger S, Tzankov A, Frank S, et al. NUT midline carcinomas and their differentials by a single molecular profiling method : a new promising diagnostic strategy illustrated by a case report. Virch Arch. 2020.
- Neumann JE, Spohn M, Obrecht D, et al. Molecular characterization of histopathological ependymoma variants. Acta Neuropathol. 2020.
- Yang D, Holsten T, Börnigen D, et al. Ependymoma relapse goes along with a relatively stable epigenome, but a severely altered tumor morphology. Brain Pathol. 2020
- Sievers P, Schrimpf D, Stichel D, et al. Posterior fossa pilocytic astrocytomas with oligodendroglial features show frequent FGFR1 activation via fusion or mutation . Acta Neuropathol. 2020.
- Hou Y, Pinheiro J, Sahm F, et al. Papillary glioneuronal tumor (PGNT) exhibits a characteristic methylation profile and fusions involving PRKCA. Acta Neuropathol. 2019.
- Capper D, Engel NW, Stichel D, et al. DNA methylation-based reclassification of olfactory neuroblastoma. Acta Neuropathol. 2018.
- Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018.
- Waszak SM, Northcott PA, Buchhalter I, et al. Spectrum and prevalence of genetic predisposition in medulloblastoma : a retrospective genetic study and prospective validation in a clinical trial cohort . Lancet Oncol. 2018 .
- Hench J, Bihl M, Bratic Hench I, et al. Satisfying your neuro-oncologist : a fast approach to routine molecular glioma diagnosis . Neuro-Oncol. 2018 .
- Sturm D, Orr BA, Toprak UH, et al. New Brain Tumor Entities Emerge from Molecular Classification of CNS-PNETs. Cell. 2016.
