Tumeurs hématolymphoides
Principaux investigateurs




Prof. Alexandar Tzankov
Leitender Arzt und Fachbereichsleiter Histopathologie und Autopsie
Pathologie
Tel. +41 61 328 68 80
Notre science
Notre groupe de recherche en hématopathologie se concentre sur les recherches translationnelles et la médecine de précision des lymphomes. Nous avons réuni une équipe de scientifiques pour faire progresser les connaissances dans les domaines distincts suivants :
Relations clonales et évolution moléculaire des lymphomes en rechute
Un thème de recherche majeur de notre groupe est le décryptage de la relation clonale et de l'évolution des lymphomes récurrents. Les rechutes des néoplasmes lymphoïdes, même après une rémission clinique de longue durée, ont été considérées jusqu'à récemment comme des excroissances directes de la tumeur primaire. Seules de rares rechutes sans lien clonal ont été documentées sous forme de rapports de cas isolés. Nous avons émis l'hypothèse que les cas de lymphomes "relapsing" clonally unrelated (véritablement "de novo") sont plus fréquents que ce que l'on pense généralement. Grâce à l'analyse comparative de la longueur des fragments de gènes de chaîne lourde d'immunoglobuline des cellules de Reed-Sternberg microdisséquées provenant d'échantillons de lymphomes de Hodgkin (LH) primaires et en rechute, nous avons pu démontrer qu'une proportion des récurrences sont clonalement sans rapport et, par conséquent, ne sont pas de véritables "rechutes" du clone malin d'origine (Clin Cancer Res. 2011;17:5268-5274). Cela peut avoir une importance clinique, car cela soulève des questions sur les thérapies cliniques agressives actuelles dans de tels cas (Figure 1).
Nous avons ensuite étendu l'hypothèse des rechutes clonally unrelated aux lymphomes diffus à grandes cellules B (DLBCL). Une cohorte de cas de DLBCL et de cas non-relapants appariés a été étudiée de manière approfondie (Leukemia 2016;30:2385-2395). Nous avons pu démontrer que de véritables récurrences de DLBCL clonally unrelated do also occur. Nous avons pu identifier deux modèles distincts d'évolution génétique dans les rechutes clonally related (early divergent/branching evolution versus late-divergent/linear progression) et de potentiels early mutational drivers of lymphomagenesis (e.g., KMT2D, MYD88, CD79B), dont certains sont désormais utilisés pour prédire la sensibilité à l'ibrutinib. En outre, nous avons pu détecter des mutations de gènes distincts qui étaient soit liées aux rechutes(BCL2, MEF2B) soit prédisaient une moindre propension aux récurrences(SOCS1). Parallèlement à ce projet, un panel de séquençage de nouvelle génération (NGS) pour les lymphomes a été développé dans notre laboratoire qui est entré dans la routine clinique après validation (Leuk Lymphoma 2018;59:1710-1716) et accréditation et qui est toujours le seul à fonctionner dans un laboratoire de diagnostic en Europe(https://www.unibas.ch/en/Research/Uni-Nova/Uni-Nova-128/Uni-Nova-128-New-treatment-concepts-for-recurrent-lymphoma.html). Pour aborder ces questions au niveau du gène unique dans le LH, nous avons établi une nouvelle technique d'enrichissement cellulaire pour une analyse génétique robuste des cellules de Reed-Sternberg (Lab Invest. 2018;98:1487-1499) et nous menons actuellement un projet correspondant.
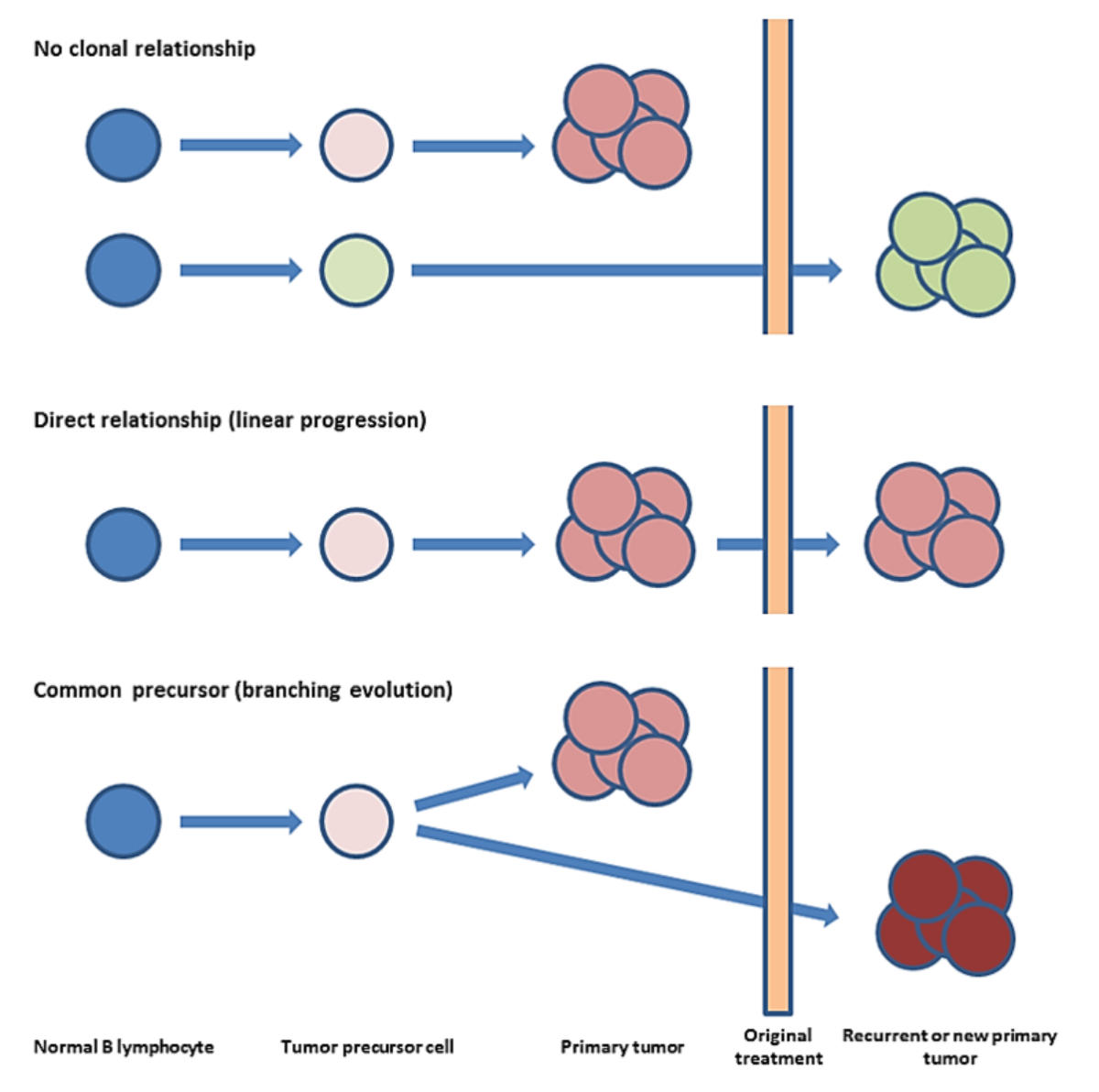
Figure 1 : Concepts des récurrences de lymphome.
Rangée supérieure : 10 à 20% des rechutes de lymphomes cliniques, c'est-à-dire la même entité tumorale apparaissant à deux moments différents, sont des secondes tumeurs sans rapport clonal. Les autres sont clonally related et présentent au moins deux modèles d'évolution moléculaire différents. Rangée du milieu : Dans le scénario d'évolution linéaire, la tumeur possède initialement de fortes mutations d'entraînement. Par conséquent, les cellules néoplastiques se développent rapidement et sans restriction, ce qui donne lieu à une tumeur primaire plutôt homogène. Une telle tumeur est quasiment éliminée par le traitement, mais une résistance acquise apparaît. Le sous-clone résistant a déjà des facteurs de croissance efficaces et réimplante la masse tumorale, ce qui donne lieu à une rechute plus rapide. Rangée inférieure : Dans le scénario de branchement, la divergence apparaît tôt dans le développement de la tumeur. La majorité des sous-populations stagnent mais un clone finit par acquérir la combinaison efficace des mutations du conducteur, se développe et donne naissance à une tumeur primaire hétérogène. La population dominante est éliminée par le traitement, mais un sous-clone intrinsèquement résistant existe et donne progressivement lieu à une rechute.
Décryptage du paysage génétique des lymphomes de la zone marginale
Nous avons décidé d'appliquer et d'étendre ce panel NGS à d'autres lymphomes orphelins, à savoir les lymphomes de la zone marginale (MZL). Il s'agit de tumeurs lymphoïdes rares comprenant trois entités différentes, mais proches du point de vue morphologique et phénotypique (LMZ splénique, nodal et extranodal). Le lymphome nodulaire est un diagnostic d'exclusion tel que défini par la classification actuelle de l'OMS, il n'a pas de phénotype définissant une maladie et ses frontières diagnostiques avec d'autres lymphomes à cellules B sont floues. Grâce à l'étude de 25 lymphomes nodulaires, nous avons identifié pour la première fois des mutations BRAF récurrentes et utiles au diagnostic, principalement V600E (Leukemia. 2018;32:2412-26 ; Figure 2). Plus important encore, la mutation V600E de ce gène est pertinente sur le plan thérapeutique, car un traitement ciblé pour cette altération moléculaire a été approuvé dans le mélanome et la leucémie à tricholeucocytes en rechute. Nous avons encore étendu nos études sur la LCM à la LCM annexielle oculaire (OA). Les données préliminaires montrent que les mutations TNFAIP3 sont très spécifiques et que les mutations BCL10 sont probablement importantes en termes de pronostic dans les CML de l'arthrose, et que les lésions lymphoïdes de l'arthrose portant des mutations composées de NF-κB et/ou des mutations de gènes codant pour l'acétyltransférase représentent très probablement des lymphomes (Der Pathologe 2019;40 Suppl. 2:S109-10). Enfin, dans les LMC du poumon, nous avons été en mesure de détecter une plus grande fréquence de cas sans mutations ponctuelles, ce qui suggère que ces tumeurs sont plus orientées vers la translocation, et nous avons démontré que - sur la base du profil mutationnel dissimilaire entre les deux entités - les DLBCL et les LMC du poumon n'ont rien à voir (Modern Pathology. 2020 ; in press).
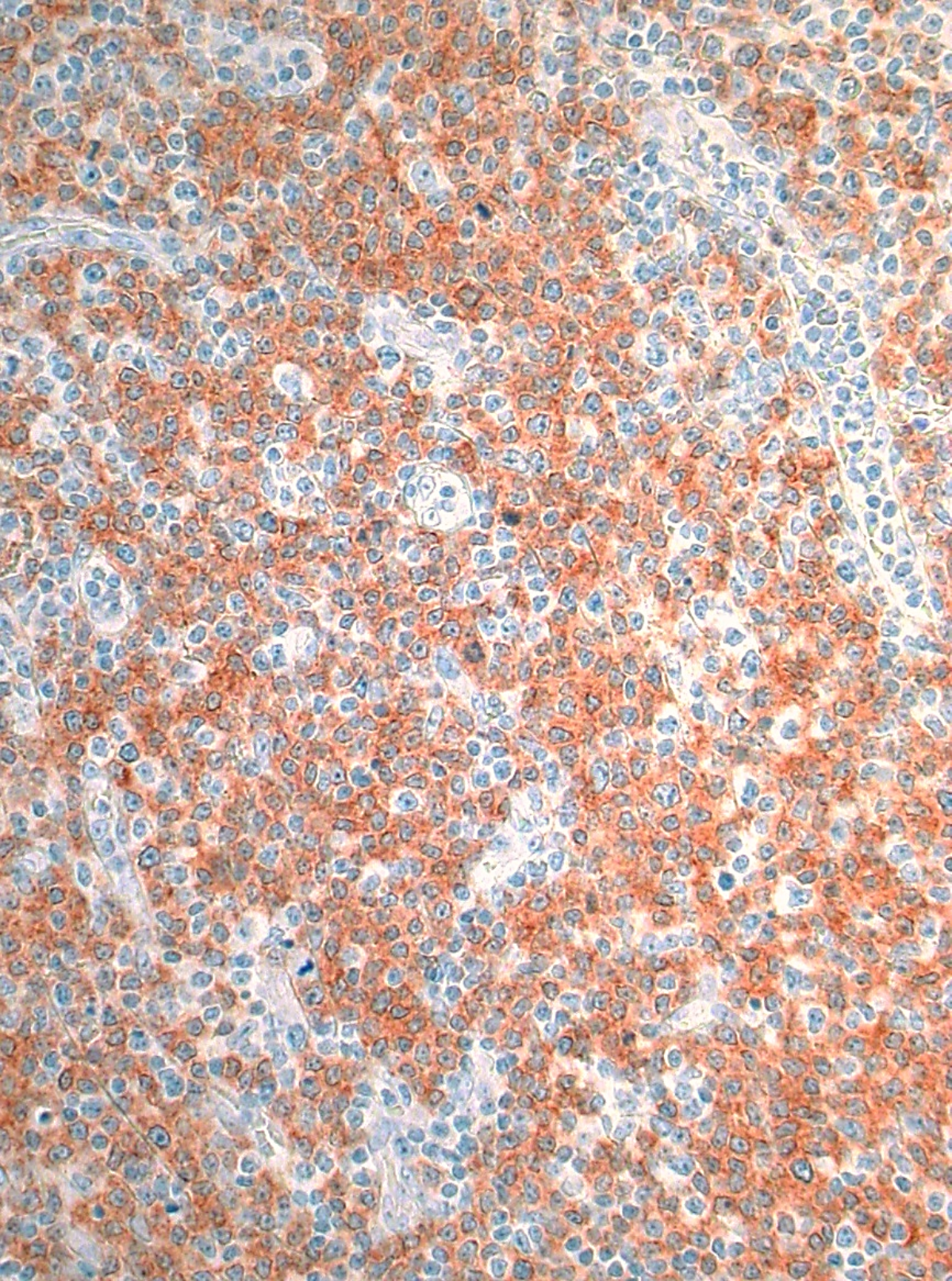
Figure 2 : Lymphome nodulaire de la zone marginale exprimant le mutant V600E de BRAF.
Une découverte importante de notre part est la détection d'une mutation récurrente dans le gène BRAF dans environ 15% des lymphomes des cellules de la zone B marginale. Cette mutation est utile dans la mesure où le produit protéique muté peut être visualisé à l'aide de méthodes de marquage simples comme illustré et est, par conséquent, applicable au diagnostic, et le produit protéique muté peut être ciblé spécifiquement, au moins dans d'autres types de cancer tels que le mélanome ou la leucémie à tricholeucocytes.
Publications sélectionnées
- Stirm K, Leary P, Bertram K, et al. Tumor cell-derived IL-10 promotes cell-autonomous growth and immune escape in diffuse large B-cell lymphoma. Oncoimmunology. 2021 Nov 22;10(1):2003533.
- Brune MM, Rau A, Overkamp M, et al. Molecular Progression of Myeloproliferative and Myelodysplastic/Myeloproliferative Neoplasms : A Study on Sequential Bone Marrow Biopsies. Cancers (Bâle). 2021 Nov 9;13(22):5605.
- Bertram K, Leary PJ, Boudesco C, et al. Inhibitors of Bcl-2 and Bruton's tyrosine kinase synergize to abrogate diffuse large B-cell lymphoma growth in vitro and in orthotopic xenotransplantation models. Leukemia. 2021 Nov 18.
- Bonfiglio F, Bruscaggin A, Guidetti F, et al. Genetic and Phenotypic Attributes of Splenic Marginal Zone Lymphoma. Blood. 2021 Oct 15:blood.2021012386.
- Drexler B, Tzankov A, Martinez M, et al. Blast counts are lower in the aspirate as compared to trephine biopsy in acute myeloid leukemia and myelodysplastic syndrome expressing CD56. Int J Lab Hematol. 2021 Mar 11.
- Vela V, Juskevicius D, Dirnhofer S, et al. paysage mutationnel des lymphomes de la zone marginale B de diverses origines : altérations organotypiques et potentiel diagnostique pour l'attribution de l'origine des organes. Virchows Arch. 2021 Sep 8.
- Brune MM, Stüssi G, Lundberg P, et al ; Groupe coopératif néerlando-belge d'hémato-oncologie (HOVON) et Groupe suisse de recherche clinique sur le cancer (SAKK). Effets du lénalidomide sur le microenvironnement de la moelle osseuse dans la leucémie myéloïde aiguë : analyse translationnelle de la cohorte HOVON103 AML/SAKK30/10 Swiss trial. Ann Hematol. 2021 Mar 2.
- Brune MM, Juskevicius D, Haslbauer J, et al. Genomic Landscape of Hodgkin Lymphoma. Cancers. 2021 Feb 8;13(4):682.
- Vela V, Juskevicius D, Prince SS, et al. Deciphering the genetic landscape of pulmonary lymphomas. Mod Pathol. 2021 Feb;34(2):371-379.
- Menter T, Tzankov A, Dirnhofer S. The tumor microenvironment of lymphomas : Insights into the potential role and modes of actions of checkpoint inhibitors . 2020 Oct 26 Hematol Oncol. doi : 10.1002/hon.2821.
- Menter T, Dirnhofer S, Tzankov A. Routine high-throughput targeted sequencing of lymphoproliferative diseases : Clinical utility and challenges. Pathologe. 2020 Dec;41(Suppl 2):143-148. French.
- Tzankov A, Duncavage E, Craig FE, et al. mastocytose. Am J Clin Pathol. 2020 Dec 14:aqaa183.
- Forte D, García-Fernández M, Sánchez-Aguilera A, et al. Bone Marrow Mesenchymal Stem Cells Support Acute Myeloid Leukemia Bioenergetics and Enhance Antioxidant Defense and Escape from Chemotherapy . Cell Metab. 2020 Nov 3;32(5):829-843.e9.
- Hebeda K, Boudova L, Beham-Schmid C, et al. Progression, transformation et manifestations inhabituelles des syndromes myélodysplasiques et des néoplasmes myélodysplasiques et myéloprolifératifs : les enseignements du XIVe cours du groupe de travail européen sur la moelle osseuse 2019. Ann Hematol. 2020 oct. 31.
- Gerlach MM, Juskevicius D, Vela V, Dirnhofer S, Tzankov A. Bone marrow infiltration of angioimmunoblastic T-cell lymphoma : identification and prognostic impact of histological patterns and diagnostic application of ancillary phenotypic and molecular analyses ; Archives of Pathology and Laboratory Medicine. 2020 144:602-611.
- Gerlach MM, Stelling-Germani A, Wu CT, Newrzela S, Döring C, Vela V, Müller A, Hartmann S, Tzankov A. Hyperméthylation du promoteur SMAD1 et absence d'expression de SMAD1 dans le lymphome de Hodgkin : une cible potentielle pour une thérapie médicamenteuse hypométhylante ; Haematologica. 2020, Apr 16.
- Menter T, Tzankov A, Zucca E, Kimby E, Hultdin M, Sundström C, Beiske K, Cogliatti S, Banz Y, Cathomas G, Karjalainen-Lindsberg ML, Grobholz R, Mazzucchelli L, Sander B, Hawle H, Hayoz S, Dirnhofer S pour le Swiss Group for Clinical Cancer Research (SAKK) et le Nordic Lymphoma Group (NLG). Prognostic implications of the microenvironment for follicular lymphoma under immunomodulation therapy ; British Journal of Haematology. 2020 189:707-717.
- Menter T, Hayoz S, Zucca E, Kimby E, Dirnhofer S, Tzankov A. Les médicaments immunomodulateurs peuvent surmonter le rôle pronostique négatif de l'axe Th17 actif dans le lymphome folliculaire : evidence from the SAKK35/10 trial ; British Journal of Haematology. 2020 190, e233-e264.
- Pillonel V, Juskevicius D, Bihl M, Stenner F, Halter JP, Dirnhofer S, Tzankov A. Routine next generation sequencing of lymphoid malignities : clinical utility and challenges from a 3-year practical experience ; Leukemia and Lymphoma. 2020, Jul 4;1-16.
- Vela V, Juskevicius D, Gerlach MM, Meyer P, Graber A, Cathomas G, Dirnhofer S, Tzankov A. High throughput sequencing reveals high specificity of TNFAIP3 mutations in ocular adnexal marginal zone B-cell lymphomas ; Hematological Oncology. 2020 38:284-292.
- Vela V, Juskevicius D, Savic Prince S, Cathomas G, Dertinger S, Diebold J, Bubendorf L, Horcic M, Singer G, Zettl A, Dirnhofer S, Tzankov A* and Menter T*. Deciphering the genetic landscape of pulmonary lymphomas ; Modern Pathology. 2020 août 27.
Earlier
- Cascione L, Rinaldi A, Bruscaggin A, Tarantelli C, Arribas AJ, Kwee I, Pecciarini L, Mensah AA, Spina V, Chung EYL, Terzi di Bergamo L, Dirnhofer S, Tzankov A, Miranda RN, Young KH, Traverse-Glehen A, Gaidano G, Swerdlow SH, Gascoyne R, Rabadan R, Ponzoni M, Bhagat G, Rossi D, Zucca E, Bertoni F.Novel insights in the genetics and epigenetics of MALT lymphoma unveiled by next generation sequencing analyses ; Haematologica. 2019 104:e558-e561
- Menter T, Lundberg P, Wenzel F, Dirks J, Fernandez P, Friess D, Dirnhofer S, Tzankov A. Les mutations RUNX1 peuvent conduire à une expression aberrante de CD79a et PAX5 dans les leucémies myélogènes aiguës : un pitfall diagnostique potentiel ; Pathobiology. 2019 86:162-166
- Hashwah H, Bertram K, Stirm K, Stelling A, Wu CT, Kasser S, Manz MG, Theocharides PA, Tzankov A, Müller A. The IL-6 signaling complex is a critical driver, negative prognostic factor and therapeutic target in diffuse large B-cell lymphoma ; European Molecular Biology Organization Molecular Medicine. 2019 11:e10576
- Juskevicius D, Jucker D, Dietsche T, Perrina V, Rufle A, Ruiz C, Dirnhofer S, Tzankov A. Novel cell enrichment technique for robust genetic analysis of archival classical Hodgkin lymphoma tissues ; Laboratory Investigation. 2018 98:1487-1499
- Juskevicius D, Müller A, Hashwah H, Lundberg P, Tzankov A, Menter T. Characterization of the mutational profile of 11 diffuse large B-cell lymphoma cell lines ; Leukemia and Lymphoma. 2018 59:1710-1716
* equal contributions
Récompenses et subventions
Collaborations
- Anne Müller, Institut de recherche sur le cancer moléculaire, Université de Zurich
- Francesco Bertoni et Davide Rossi, Institute of Oncology Research, Bellinzona
- Juerg Schwaller, Département de biomédecine, Université de Bâle
- Ken H Young, Université de Duke, Caroline du Nord
- Radek Skoda, Département de biomédecine, Université de Bâle
