Chirurgie viscérale et médecine de précision
Notre science
La médecine de précision consiste à administrer le bon traitement aux patients au bon moment. En prenant le cancer rectal comme exemple, les patients atteints d'un cancer rectal de stade intermédiaire sont généralement traités par chimioradiothérapie, suivie d'une excision chirurgicale pour éliminer toute maladie résiduelle. L'analyse histologique post-chirurgicale révèle que ~25% des patients obtiennent une réponse complète à la chimioradiothérapie, ce qui signifie qu'ils n'auraient pas eu besoin d'une intervention chirurgicale. Pour les patients restants qui n'obtiennent pas une réponse complète à la chimioradiothérapie, des options thérapeutiques alternatives sont souhaitables. Il est donc urgent d'identifier des biomarqueurs robustes qui permettront de prédire avec précision quels patients obtiendront probablement une bonne réponse à la chimioradiothérapie, leur évitant ainsi une opération chirurgicale inutile. D'autre part, des systèmes modèles qui reflètent le comportement biologique des tumeurs chez les patients individuels afin de tester des options thérapeutiques alternatives de manière ex vivo sont essentiels. Notre laboratoire adopte plusieurs approches complémentaires pour relever les défis de l'oncologie de précision.
Modèles ex vivo pour le test de médicaments
Les organoïdes dérivés de patients (PDO) ont montré qu'ils conservaient les caractéristiques moléculaires des tumeurs d'origine et qu'ils ressemblaient mieux à l'hétérogénéité tumorale que les méthodes traditionnelles de culture cellulaire en deux dimensions à partir de clones de cellules uniques. Elles sont donc souvent utilisées comme modèles précliniques ex vivo pour la prédiction de la réponse aux médicaments. En collaboration avec les chirurgiens viscéraux de Clarunis, nous avons initié la collecte prospective de tumeurs de patients afin d'établir une biobanque vivante de PDO. A ce jour, nous avons établi avec succès une collection de ~100 PDOs (Fig. 1a). Nous avons également mis en place un protocole pour tester les médicaments et surveiller la réponse en temps réel pendant cinq jours. En utilisant ce protocole, nous avons montré que la 6-mercaptopurine réduit efficacement la croissance cellulaire et induit l'apoptose dans les tumeurs colorectales avec surexpression de l'ADSL, un gène ayant un rôle critique dans la synthèse de la purine.
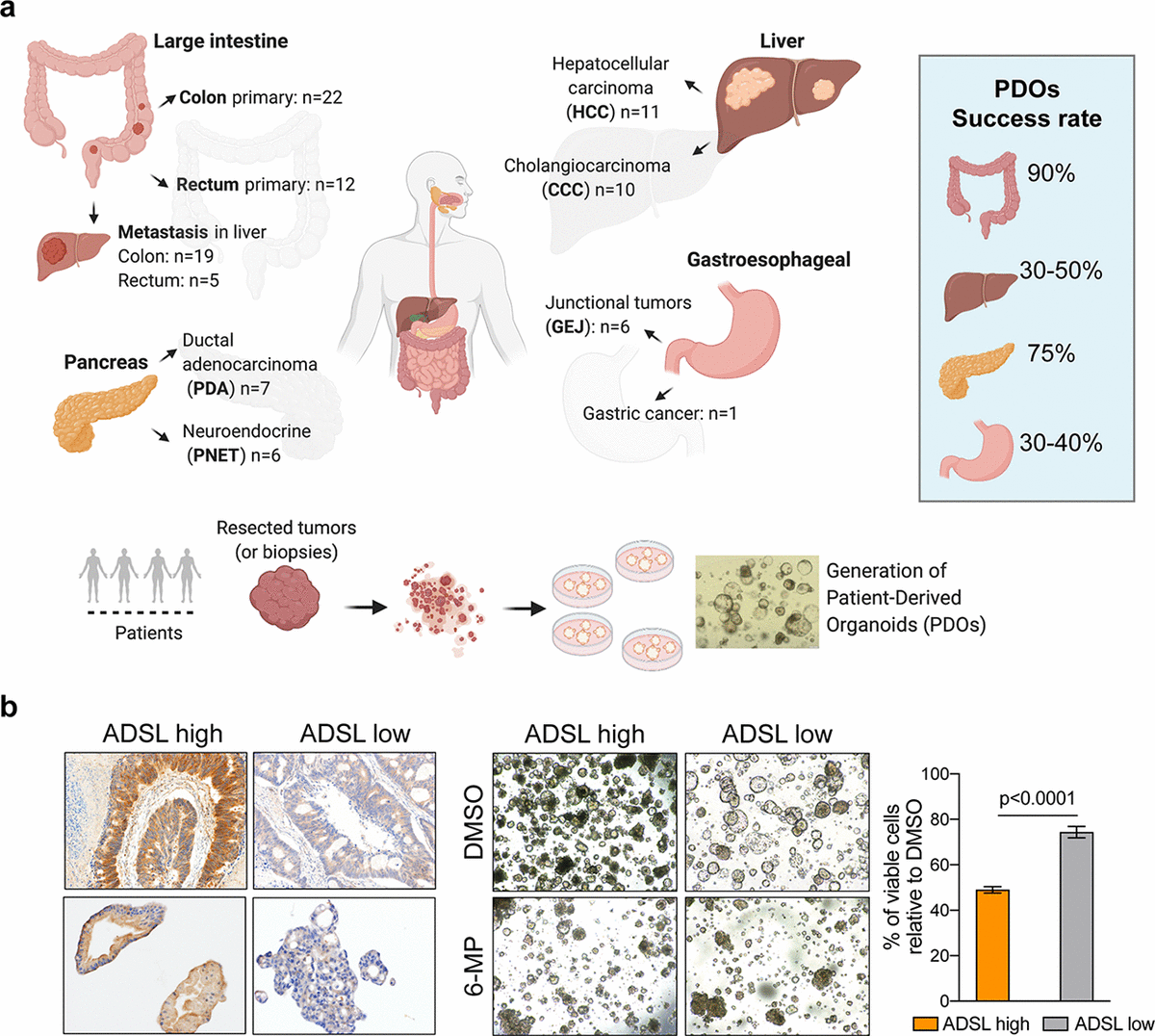
Fig. 1 : (a) Nous avons optimisé les protocoles pour dériver des organoïdes dérivés de patients à partir de spécimens de tissus. En collaboration avec les chirurgiens de Clarunis, nous avons établi ~100 organoïdes dans notre biobanque vivante. (b) Notre protocole de dépistage des médicaments dans les PDO nous a permis de démontrer que la surexpression de l'ADSL est un biomarqueur de la réponse à la 6-mercaptopurine dans les tumeurs colorectales.
Découverte de vulnérabilités cancéreuses
Le concept de la létalité synthétique a permis d'étendre l'oncologie de précision afin de permettre le ciblage de gènes non-médicables en perturbant leurs interacteurs génétiques. Par exemple, GATA3 est muté dans 15 à 18% des cancers du sein à récepteurs d'œstrogènes positifs, mais n'est pas ciblable. En interrogeant systématiquement un large éventail d'ARNi, nous avons identifié MDM2 comme l'un des principaux interacteurs synthétiques létaux de GATA3. Nous avons constaté que l'inhibition pharmacologique de MDM2 par l'idasanutline réduit la prolifération cellulaire et induit l'apoptose dans les cancers déficients en GATA3 in vitro et dans deux modèles indépendants in vivo (zebrafish et chicken chorioallantoic membrane models). Les PDO mutants de GATA3 sont également plus sensibles à l'idasanutline que les PDO de type sauvage de GATA3. Avec l'idasanutline et d'autres inhibiteurs de MDM2 largement disponibles, nos résultats peuvent être rapidement traduits en essais cliniques pour évaluer l'efficacité chez le patient et le statut de mutation GATA3 en tant que biomarqueur prédictif de la réponse à l'idasanutline dans le cancer du sein.
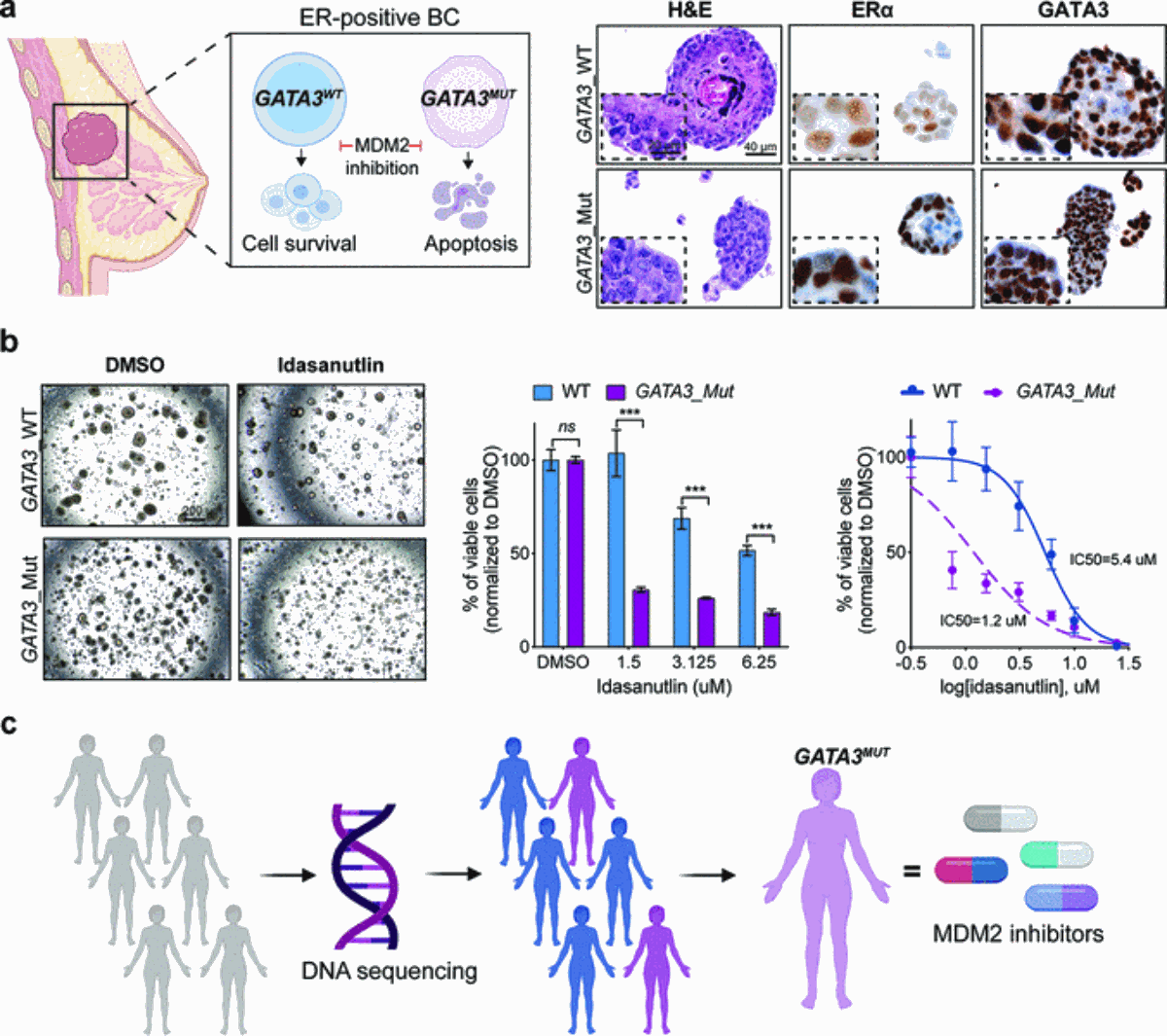
Fig. 2 : (a) MDM2 est une nouvelle cible létale synthétique dans le cancer du sein GATA3-mutant avec récepteur d'œstrogènes (ER) positif. Organoïdes dérivés de cancers du sein GATA3 sauvages (WT) et mutants GATA3. (b) Les mutations GATA3 sensibilisent les PDO au traitement par l'idasanutline. (c) Stratégie de dépistage génétique pour permettre un ciblage précis du cancer du sein mutant GATA3.
L'ADN sans cellule comme substitut tumoral peu invasif
La médecine de précision n'est possible qu'avec des matériaux tumoraux, et ceux qui peuvent être obtenus par des procédures peu invasives (par ex. prélèvement de sang) sont particulièrement souhaitables. Il a été démontré que l'ADN libre de cellules dérivé du plasma (cfDNA) reflète la composition génétique des tumeurs et peut servir de substitut tumoral pour la détection des mutations et le suivi de la maladie pendant le traitement. Le cfDNA peut être particulièrement utile en tant que substitut génétique pour les tumeurs difficiles ou impossibles à biopsier. Ceci est important pour le carcinome hépatocellulaire (CHC) car son diagnostic ne nécessite pas toujours une biopsie tissulaire ; les matériaux tissulaires tumoraux chez les patients inopérables sont généralement indisponibles. Le manque de matériel tumoral empêche l'adoption plus large de la médecine de précision dans le CHC. Nous avons constaté que le profilage génétique de l'ADNc dérivé du plasma était un substitut adéquat du CHC primaire chez les patients atteints d'une tumeur de grande taille ou d'une maladie métastatique (publication récompensée par le prix 2019 de la Swiss Foundation against Liver Cancer). Nous sommes maintenant en train d'étudier si le cfDNA peut aider à surveiller la maladie dans le CHC.
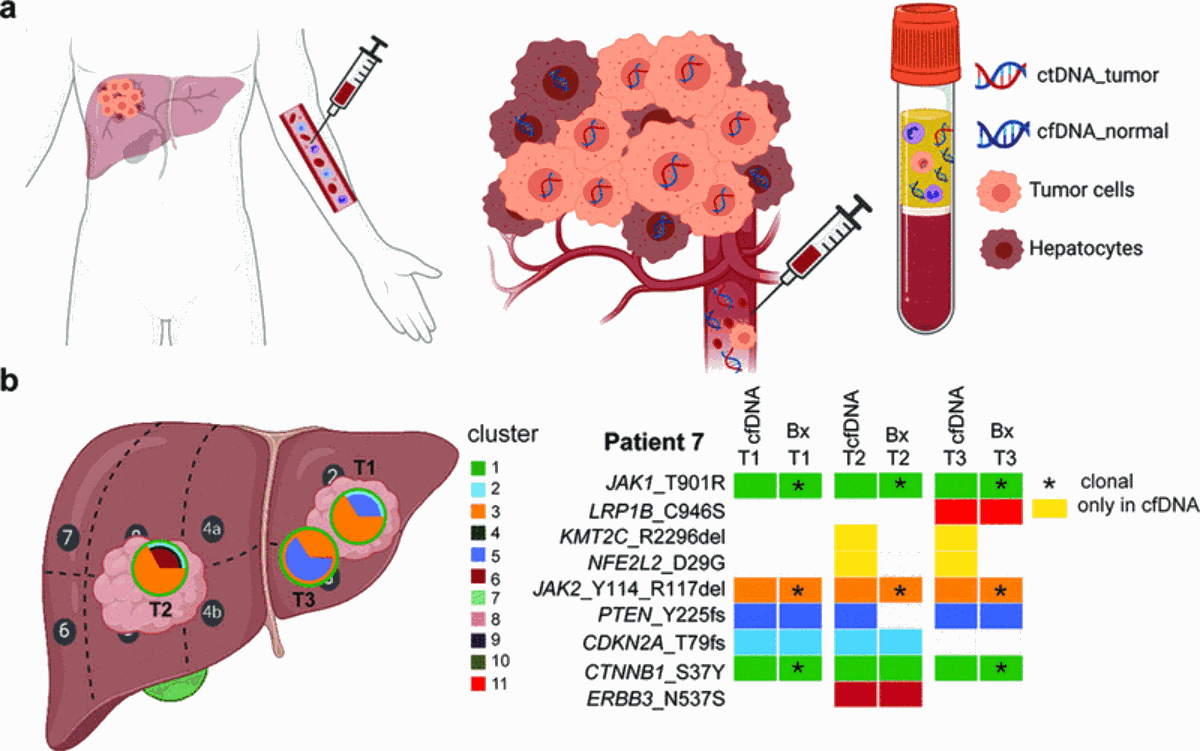
Fig. 3 : (a) ADN libre de cellules dérivé du plasma comme substitut tumoral pour la détection des mutations. (b) Collecte longitudinale d'ADN libre de cellules en tant qu'outil de surveillance de la maladie dans le carcinome hépatocellulaire.
Publications sélectionnées
-
Ghosh, S. et al. Prevention of dsRNA-induced interferon signaling by AGO1x is linked to breast cancer cell proliferation. EMBO J. e103922 (2020) doi:10.15252/embj.2019103922.
-
Cyrta, J. et al. Role of specialized composition of SWI/SNF complexes in prostate cancer lineage plasticity. Nat. Commun. 11, 5549 (2020).
-
Ng, C. K. Y. et al. Genetic profiling using plasma-derived cell-free DNA in therapy-naïve hepatocellular carcinoma patients : a pilot study. Ann. Oncol. 29, 1286-1291 (2018).
-
Bianco, G. et al. GATA3 and MDM2 are synthetic lethal in estrogen receptor-positive breast cancers. Cancer Biology 564 (2020). (Pré-impression)
-
Montazeri, H. et al. Systematic Identification of Novel Cancer Genes through Analysis of Deep shRNA Perturbation Screens. Génomique 305 (2019). (Pre-print)
-
Bertucci, F. et al. Genomic characterization of metastatic breast cancers. Nature 569, 560-564 (2019).
La liste complète des publications peut être consultée sur https://pubmed.ncbi.nlm.nih.gov/?term=Piscuoglio&sort=date&size=200
Récompenses et subventions
